Load CMIP6 Data with Intake ESM¶
This notebook demonstrates how to access Google Cloud CMIP6 data using intake-esm.
Loading a catalog¶
import warnings
warnings.filterwarnings("ignore")
import intake
url = "https://storage.googleapis.com/cmip6/pangeo-cmip6.json"
col = intake.open_esm_datastore(url)
col
Matplotlib is building the font cache; this may take a moment.
pangeo-cmip6 catalog with 7483 dataset(s) from 512699 asset(s):
| unique | |
|---|---|
| activity_id | 18 |
| institution_id | 37 |
| source_id | 87 |
| experiment_id | 172 |
| member_id | 651 |
| table_id | 38 |
| variable_id | 710 |
| grid_label | 11 |
| zstore | 512699 |
| dcpp_init_year | 60 |
| version | 684 |
The summary above tells us that this catalog contains over 268,000 data assets. We can get more information on the individual data assets contained in the catalog by calling the underlying dataframe created when it is initialized:
Catalog Contents¶
col.df.head()
| activity_id | institution_id | source_id | experiment_id | member_id | table_id | variable_id | grid_label | zstore | dcpp_init_year | version | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | HighResMIP | CMCC | CMCC-CM2-HR4 | highresSST-present | r1i1p1f1 | Amon | hus | gn | gs://cmip6/CMIP6/HighResMIP/CMCC/CMCC-CM2-HR4/... | NaN | 20170706 |
| 1 | HighResMIP | CMCC | CMCC-CM2-HR4 | highresSST-present | r1i1p1f1 | Amon | rsdt | gn | gs://cmip6/CMIP6/HighResMIP/CMCC/CMCC-CM2-HR4/... | NaN | 20170706 |
| 2 | HighResMIP | CMCC | CMCC-CM2-HR4 | highresSST-present | r1i1p1f1 | Amon | prw | gn | gs://cmip6/CMIP6/HighResMIP/CMCC/CMCC-CM2-HR4/... | NaN | 20170706 |
| 3 | HighResMIP | CMCC | CMCC-CM2-HR4 | highresSST-present | r1i1p1f1 | Amon | rlus | gn | gs://cmip6/CMIP6/HighResMIP/CMCC/CMCC-CM2-HR4/... | NaN | 20170706 |
| 4 | HighResMIP | CMCC | CMCC-CM2-HR4 | highresSST-present | r1i1p1f1 | Amon | rlds | gn | gs://cmip6/CMIP6/HighResMIP/CMCC/CMCC-CM2-HR4/... | NaN | 20170706 |
The first data asset listed in the catalog contains:
the ambient aerosol optical thickness at 550nm (
variable_id='od550aer'), as a function of latitude, longitude, time,in an individual climate model experiment with the Taiwan Earth System Model 1.0 model (
source_id='TaiESM1'),forced by the Historical transient with SSTs prescribed from historical experiment (
experiment_id='histSST'),developed by the Taiwan Research Center for Environmental Changes (
instution_id='AS-RCEC'),run as part of the Aerosols and Chemistry Model Intercomparison Project (
activity_id='AerChemMIP')
And is located in Google Cloud Storage at
gs://cmip6/AerChemMIP/AS-RCEC/TaiESM1/histSST/r1i1p1f1/AERmon/od550aer/gn/.
Finding unique entries¶
Let’s query the data to see what models (source_id), experiments
(experiment_id) and temporal frequencies (table_id) are available.
import pprint
uni_dict = col.unique(["source_id", "experiment_id", "table_id"])
pprint.pprint(uni_dict, compact=True)
{'experiment_id': {'count': 172,
'values': ['histSST-piNTCF', 'abrupt-solm4p',
'piClim-2xdust', 'aqua-p4K-lwoff', 'esm-hist',
'r7i1p1f1', 'dcppC-amv-Trop-neg',
'faf-heat-NA50pct', 'ssp245-covid',
'ssp245-cov-strgreen', 'histSST-1950HC',
'dcppC-pac-pacemaker', 'ssp370SST-lowCH4',
'piClim-histghg', 'ssp119', 'dcppA-hindcast',
'abrupt-solp4p', 'ssp460', 'ssp370SST-ssp126Lu',
'piClim-NTCF', 'hist-resIPO', 'aqua-4xCO2',
'piClim-anthro', 'ssp585-bgc', 'r4i1p1f1',
'esm-piControl', 'dcppC-amv-ExTrop-neg',
'esm-pi-cdr-pulse', 'control-1950',
'pdSST-pdSICSIT', 'faf-stress', 'rcp45-cmip5',
'hist-1950HC', 'lgm', 'ssp370-lowNTCF',
'piClim-O3', 'faf-passiveheat', 'ssp245-GHG',
'histSST-piAer', 'abrupt-0p5xCO2', 'faf-heat',
'hist-totalO3', 'piClim-lu', 'piClim-2xfire',
'aqua-p4K', 'piClim-BC', 'piClim-NOx',
'piClim-ghg', 'dcppC-pac-control', 'hist-piAer',
'pa-piAntSIC', 'abrupt-4xCO2', 'piClim-aer',
'dcppC-ipv-NexTrop-neg', 'land-hist-altStartYear',
'pdSST-piArcSIC', 'lig127k', 'midHolocene',
'highresSST-present', 'piClim-histaer',
'dcppC-amv-ExTrop-pos', 'amip-4xCO2',
'aqua-control', 'piClim-histnat',
'ssp370-ssp126Lu', 'hist-bgc',
'dcppC-amv-Trop-pos', 'pdSST-pdSIC',
'1pctCO2-rad', 'dcppC-hindcast-noElChichon',
'amip-p4K', 'esm-pi-CO2pulse', 'dcppC-ipv-pos',
'piControl-spinup', 'ssp245-cov-modgreen',
'esm-ssp585', 'histSST-piCH4', 'hist-CO2',
'land-hist', 'piControl', 'histSST-piO3',
'pdSST-piAntSIC', 'pdSST-futArcSICSIT',
'ssp245-cov-fossil', 'piClim-4xCO2',
'abrupt-2xCO2', '1pctCO2-bgc', 'piClim-control',
'aqua-control-lwoff', 'futSST-pdSIC',
'piClim-SO2', 'hist-1950', 'hist-volc',
'past1000', 'ssp370', 'amip-hist',
'pdSST-futArcSIC', 'historical-cmip5',
'dcppC-hindcast-noPinatubo', 'piClim-OC',
'amip-future4K', 'hist-aer', 'pa-pdSIC',
'ssp370SST', 'dcppC-ipv-NexTrop-pos',
'esm-ssp585-ssp126Lu', 'pa-futAntSIC',
'piClim-HC', 'dcppC-amv-neg', 'ssp585',
'ssp534-over', 'dcppA-assim', 'faf-heat-NA0pct',
'piClim-VOC', 'land-noLu', 'deforest-globe',
'piClim-N2O', 'dcppC-amv-pos', 'pdSST-futAntSIC',
'ssp126-ssp370Lu', 'piClim-CH4', 'dcppC-ipv-neg',
'hist-piNTCF', 'r5i1p1f1', 'pdSST-futOkhotskSIC',
'histSST', 'pdSST-futBKSeasSIC',
'esm-piControl-spinup', 'piClim-2xDMS',
'ssp370SST-lowNTCF', 'ssp126', 'ssp370pdSST',
'historical', 'dcppC-hindcast-noAgung',
'pa-futArcSIC', 'r6i1p1f1', 'piSST-piSIC',
'rcp85-cmip5', 'ssp434', 'piClim-histall',
'faf-water', 'ssp245', 'dcppC-atl-control',
'hist-sol', 'historical-ext', 'piClim-2xNOx',
'hist-nat-cmip5', 'piSST-pdSIC', 'piClim-2xss',
'hist-nat', 'piControl-cmip5', 'pa-piArcSIC',
'highresSST-future', 'hist-noLu', 'amip-lwoff',
'hist-stratO3', '1pctCO2', 'hist-aer-cmip5',
'amip', 'hist-GHG', 'ssp245-aer', 'amip-m4K',
'dcppC-atl-pacemaker', 'amip-p4K-lwoff',
'rcp26-cmip5', '1pctCO2-cdr', 'omip1', 'faf-all',
'ssp245-nat', 'hist-GHG-cmip5', 'ssp245-stratO3',
'piClim-2xVOC']},
'source_id': {'count': 87,
'values': ['ECMWF-IFS-LR', 'EC-Earth3P-VHR', 'UKESM1-0-LL',
'CAMS-CSM1-0', 'EC-Earth3', 'CNRM-CM6-1-HR',
'CMCC-CM2-HR4', 'EC-Earth3-AerChem', 'CESM2-FV2',
'CNRM-ESM2-1', 'GFDL-CM4C192', 'INM-CM4-8',
'AWI-ESM-1-1-LR', 'CAS-ESM2-0', 'GFDL-ESM4',
'CMCC-CM2-SR5', 'MIROC-ES2H', 'FGOALS-g3',
'GISS-E2-1-G-CC', 'MRI-AGCM3-2-H', 'TaiESM1',
'GISS-E2-1-H', 'CMCC-CM2-VHR4', 'CESM2-WACCM',
'MPI-ESM1-2-LR', 'HadGEM3-GC31-LL', 'CanESM5',
'HadGEM3-GC31-HM', 'AWI-CM-1-1-MR', 'CanESM5-CanOE',
'MPI-ESM1-2-XR', 'BCC-CSM2-MR', 'EC-Earth3-Veg',
'FIO-ESM-2-0', 'E3SM-1-1-ECA', 'MPI-ESM1-2-HR',
'CESM2', 'ACCESS-CM2', 'EC-Earth3-CC', 'NESM3',
'CESM1-1-CAM5-CMIP5', 'MRI-AGCM3-2-S', 'ECMWF-IFS-HR',
'BCC-ESM1', 'NorCPM1', 'EC-Earth3P-HR', 'CNRM-CM6-1',
'KIOST-ESM', 'FGOALS-f3-H', 'NorESM1-F',
'GISS-E2-1-G', 'IPSL-CM5A2-INCA', 'IPSL-CM6A-LR',
'INM-CM5-H', 'NorESM2-MM', 'CIESM', 'CESM1-WACCM-SC',
'SAM0-UNICON', 'HadGEM3-GC31-MM', 'ssp585', 'MIROC6',
'IPSL-CM6A-ATM-HR', 'ACCESS-ESM1-5',
'CESM2-WACCM-FV2', 'MPI-ESM-1-2-HAM', 'GFDL-CM4',
'HadGEM3-GC31-LM', 'EC-Earth3P', 'MCM-UA-1-0',
'GFDL-OM4p5B', 'GFDL-ESM2M', 'EC-Earth3-LR',
'EC-Earth3-Veg-LR', 'BCC-CSM2-HR', 'GFDL-AM4',
'FGOALS-f3-L', 'E3SM-1-0', 'CMCC-ESM2', 'E3SM-1-1',
'KACE-1-0-G', 'IITM-ESM', 'IPSL-CM6A-LR-INCA',
'MIROC-ES2L', 'GISS-E2-2-G', 'INM-CM5-0',
'NorESM2-LM', 'MRI-ESM2-0']},
'table_id': {'count': 38,
'values': ['EdayZ', 'AERmon', 'CF3hr', 'hus', 'IfxGre', 'Lmon',
'3hr', 'CFmon', 'Efx', 'SIclim', '6hrPlevPt', 'Eclim',
'Emon', 'Omon', 'Ofx', 'Odec', 'E1hrClimMon', 'fx',
'ImonGre', 'AERmonZ', 'Amon', 'AERday', 'day', 'CFday',
'Eday', 'Oclim', '6hrLev', 'Eyr', 'Aclim', 'E3hr',
'SImon', 'AERhr', 'LImon', 'SIday', '6hrPlev', 'Oday',
'Oyr', 'EmonZ']}}
Searching for specific datasets¶
In the example below, we are are going to search for the following:
variables:
o2which stands formole_concentration_of_dissolved_molecular_oxygen_in_sea_waterexperiments:
['historical', 'ssp585']:historical: all forcing of the recent past.ssp585: emission-driven RCP8.5 based on SSP5.
table_id:
Oyrwhich stands for annual mean variables on the ocean grid.grid_label:
gnwhich stands for data reported on a model’s native grid.
For more details on the CMIP6 vocabulary, please check this website, and Core Controlled Vocabularies (CVs) for use in CMIP6 GitHub repository.
cat = col.search(
experiment_id=["historical", "ssp585"],
table_id="Oyr",
variable_id="o2",
grid_label="gn",
)
cat
pangeo-cmip6 catalog with 28 dataset(s) from 180 asset(s):
| unique | |
|---|---|
| activity_id | 2 |
| institution_id | 13 |
| source_id | 15 |
| experiment_id | 2 |
| member_id | 47 |
| table_id | 1 |
| variable_id | 1 |
| grid_label | 1 |
| zstore | 180 |
| dcpp_init_year | 0 |
| version | 31 |
cat.df.head()
| activity_id | institution_id | source_id | experiment_id | member_id | table_id | variable_id | grid_label | zstore | dcpp_init_year | version | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CMIP | IPSL | IPSL-CM6A-LR | historical | r12i1p1f1 | Oyr | o2 | gn | gs://cmip6/CMIP6/CMIP/IPSL/IPSL-CM6A-LR/histor... | NaN | 20180803 |
| 1 | CMIP | IPSL | IPSL-CM6A-LR | historical | r21i1p1f1 | Oyr | o2 | gn | gs://cmip6/CMIP6/CMIP/IPSL/IPSL-CM6A-LR/histor... | NaN | 20180803 |
| 2 | CMIP | IPSL | IPSL-CM6A-LR | historical | r11i1p1f1 | Oyr | o2 | gn | gs://cmip6/CMIP6/CMIP/IPSL/IPSL-CM6A-LR/histor... | NaN | 20180803 |
| 3 | CMIP | IPSL | IPSL-CM6A-LR | historical | r10i1p1f1 | Oyr | o2 | gn | gs://cmip6/CMIP6/CMIP/IPSL/IPSL-CM6A-LR/histor... | NaN | 20180803 |
| 4 | CMIP | IPSL | IPSL-CM6A-LR | historical | r1i1p1f1 | Oyr | o2 | gn | gs://cmip6/CMIP6/CMIP/IPSL/IPSL-CM6A-LR/histor... | NaN | 20180803 |
Loading datasets Using to_dataset_dict()¶
dset_dict = cat.to_dataset_dict(
zarr_kwargs={"consolidated": True, "decode_times": True, "use_cftime": True}
)
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'
[key for key in dset_dict.keys()]
['ScenarioMIP.NCC.NorESM2-MM.ssp585.Oyr.gn',
'CMIP.MRI.MRI-ESM2-0.historical.Oyr.gn',
'ScenarioMIP.IPSL.IPSL-CM6A-LR.ssp585.Oyr.gn',
'ScenarioMIP.CMCC.CMCC-ESM2.ssp585.Oyr.gn',
'ScenarioMIP.EC-Earth-Consortium.EC-Earth3-CC.ssp585.Oyr.gn',
'ScenarioMIP.MIROC.MIROC-ES2L.ssp585.Oyr.gn',
'CMIP.MPI-M.MPI-ESM1-2-HR.historical.Oyr.gn',
'ScenarioMIP.DWD.MPI-ESM1-2-HR.ssp585.Oyr.gn',
'CMIP.CCCma.CanESM5-CanOE.historical.Oyr.gn',
'ScenarioMIP.NCAR.CESM2.ssp585.Oyr.gn',
'ScenarioMIP.NCC.NorESM2-LM.ssp585.Oyr.gn',
'CMIP.CCCma.CanESM5.historical.Oyr.gn',
'CMIP.EC-Earth-Consortium.EC-Earth3-CC.historical.Oyr.gn',
'CMIP.NCC.NorESM2-MM.historical.Oyr.gn',
'ScenarioMIP.MPI-M.MPI-ESM1-2-LR.ssp585.Oyr.gn',
'CMIP.CMCC.CMCC-ESM2.historical.Oyr.gn',
'CMIP.CSIRO.ACCESS-ESM1-5.historical.Oyr.gn',
'CMIP.MIROC.MIROC-ES2L.historical.Oyr.gn',
'CMIP.HAMMOZ-Consortium.MPI-ESM-1-2-HAM.historical.Oyr.gn',
'ScenarioMIP.CSIRO.ACCESS-ESM1-5.ssp585.Oyr.gn',
'CMIP.MPI-M.MPI-ESM1-2-LR.historical.Oyr.gn',
'CMIP.IPSL.IPSL-CM5A2-INCA.historical.Oyr.gn',
'ScenarioMIP.CCCma.CanESM5.ssp585.Oyr.gn',
'ScenarioMIP.DKRZ.MPI-ESM1-2-HR.ssp585.Oyr.gn',
'CMIP.NCC.NorESM2-LM.historical.Oyr.gn',
'CMIP.IPSL.IPSL-CM6A-LR.historical.Oyr.gn',
'ScenarioMIP.MRI.MRI-ESM2-0.ssp585.Oyr.gn',
'ScenarioMIP.CCCma.CanESM5-CanOE.ssp585.Oyr.gn']
We can access a particular dataset as follows:
ds = dset_dict["CMIP.CCCma.CanESM5.historical.Oyr.gn"]
print(ds)
<xarray.Dataset>
Dimensions: (i: 360, j: 291, lev: 45, bnds: 2, member_id: 35, time: 165, vertices: 4)
Coordinates:
* i (i) int32 0 1 2 3 4 5 6 ... 353 354 355 356 357 358 359
* j (j) int32 0 1 2 3 4 5 6 ... 284 285 286 287 288 289 290
latitude (j, i) float64 dask.array<chunksize=(291, 360), meta=np.ndarray>
* lev (lev) float64 3.047 9.454 16.36 ... 5.375e+03 5.625e+03
lev_bnds (lev, bnds) float64 dask.array<chunksize=(45, 2), meta=np.ndarray>
longitude (j, i) float64 dask.array<chunksize=(291, 360), meta=np.ndarray>
* time (time) object 1850-07-02 12:00:00 ... 2014-07-02 12:0...
time_bnds (time, bnds) object dask.array<chunksize=(165, 2), meta=np.ndarray>
* member_id (member_id) <U9 'r24i1p1f1' 'r16i1p1f1' ... 'r10i1p1f1'
Dimensions without coordinates: bnds, vertices
Data variables:
o2 (member_id, time, lev, j, i) float32 dask.array<chunksize=(1, 12, 45, 291, 360), meta=np.ndarray>
vertices_latitude (j, i, vertices) float64 dask.array<chunksize=(291, 360, 4), meta=np.ndarray>
vertices_longitude (j, i, vertices) float64 dask.array<chunksize=(291, 360, 4), meta=np.ndarray>
Attributes: (12/58)
source_id: CanESM5
branch_time_in_child: 0.0
contact: ec.cccma.info-info.ccmac.ec@canada.ca
parent_activity_id: CMIP
CCCma_runid: rc3.1-his10
references: Geophysical Model Development Special issue ...
... ...
table_info: Creation Date:(20 February 2019) MD5:374fbe5...
CCCma_pycmor_hash: 33c30511acc319a98240633965a04ca99c26427e
status: 2019-10-25;created;by nhn2@columbia.edu
parent_mip_era: CMIP6
sub_experiment_id: none
intake_esm_dataset_key: CMIP.CCCma.CanESM5.historical.Oyr.gn
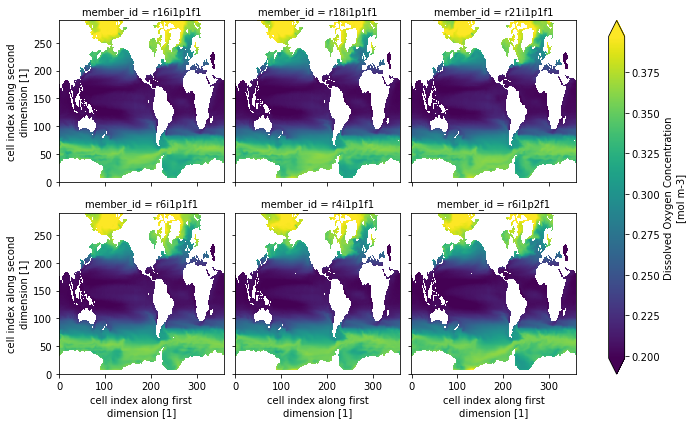
Let’s create a quick plot for a slice of the data:
ds.o2.isel(time=0, lev=0, member_id=range(1, 24, 4)).plot(col="member_id", col_wrap=3, robust=True)
<xarray.plot.facetgrid.FacetGrid at 0x7efd93bfb610>
Using custom preprocessing functions¶
When comparing many models it is often necessary to preprocess (e.g. rename
certain variables) them before running some analysis step. The preprocess
argument lets the user pass a function, which is executed for each loaded asset
before aggregations.
cat_pp = col.search(
experiment_id=["historical"],
table_id="Oyr",
variable_id="o2",
grid_label="gn",
source_id=["IPSL-CM6A-LR", "CanESM5"],
member_id="r10i1p1f1",
)
cat_pp.df
| activity_id | institution_id | source_id | experiment_id | member_id | table_id | variable_id | grid_label | zstore | dcpp_init_year | version | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CMIP | IPSL | IPSL-CM6A-LR | historical | r10i1p1f1 | Oyr | o2 | gn | gs://cmip6/CMIP6/CMIP/IPSL/IPSL-CM6A-LR/histor... | NaN | 20180803 |
| 1 | CMIP | CCCma | CanESM5 | historical | r10i1p1f1 | Oyr | o2 | gn | gs://cmip6/CMIP6/CMIP/CCCma/CanESM5/historical... | NaN | 20190429 |
# load the example
dset_dict_raw = cat_pp.to_dataset_dict(zarr_kwargs={"consolidated": True})
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'
for k, ds in dset_dict_raw.items():
print(f"dataset key={k}\n\tdimensions={sorted(list(ds.dims))}\n")
dataset key=CMIP.IPSL.IPSL-CM6A-LR.historical.Oyr.gn
dimensions=['axis_nbounds', 'member_id', 'nvertex', 'olevel', 'time', 'x', 'y']
dataset key=CMIP.CCCma.CanESM5.historical.Oyr.gn
dimensions=['bnds', 'i', 'j', 'lev', 'member_id', 'time', 'vertices']
Note
Note that both models follow a different naming scheme. We can define a little
helper function and pass it to .to_dataset_dict() to fix this. For
demonstration purposes we will focus on the vertical level dimension which is
called lev in CanESM5 and olevel in IPSL-CM6A-LR.
def helper_func(ds):
"""Rename `olevel` dim to `lev`"""
ds = ds.copy()
# a short example
if "olevel" in ds.dims:
ds = ds.rename({"olevel": "lev"})
return ds
dset_dict_fixed = cat_pp.to_dataset_dict(zarr_kwargs={"consolidated": True}, preprocess=helper_func)
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'
for k, ds in dset_dict_fixed.items():
print(f"dataset key={k}\n\tdimensions={sorted(list(ds.dims))}\n")
dataset key=CMIP.IPSL.IPSL-CM6A-LR.historical.Oyr.gn
dimensions=['axis_nbounds', 'lev', 'member_id', 'nvertex', 'time', 'x', 'y']
dataset key=CMIP.CCCma.CanESM5.historical.Oyr.gn
dimensions=['bnds', 'i', 'j', 'lev', 'member_id', 'time', 'vertices']
This was just an example for one dimension.
Note
Check out cmip6-preprocessing package for a full renaming function for all available CMIP6 models and some other utilities.
import intake_esm # just to display version information
intake_esm.show_versions()
INSTALLED VERSIONS
------------------
cftime: 1.5.0
dask: 2021.08.0
fastprogress: 0.2.7
fsspec: 2021.07.0
gcsfs: 2021.07.0
intake: 0.6.3
intake_esm: 0.0.0
netCDF4: 1.5.7
pandas: 1.3.2
requests: 2.26.0
s3fs: 2021.07.0
xarray: 0.19.0
zarr: 2.8.3
